Home
Products
KU-0063794
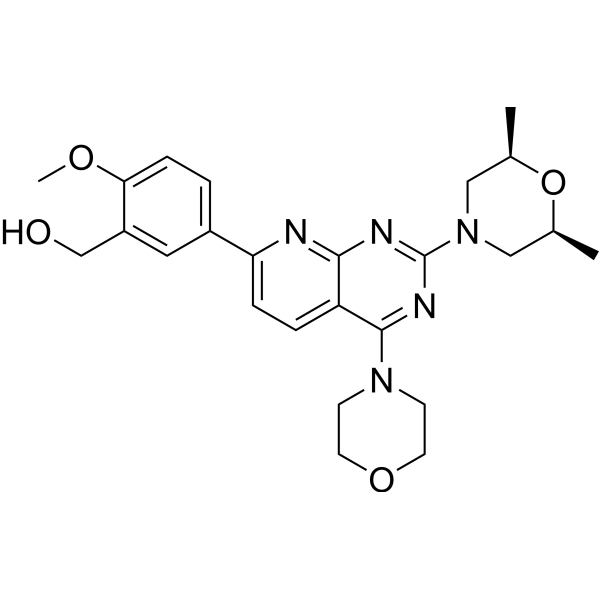


| Product Name | KU-0063794 |
| Price: | Inquiry |
| Catalog No.: | CN00328 |
| CAS No.: | 938440-64-3 |
| Molecular Formula: | C25H31N5O4 |
| Molecular Weight: | 465.54 g/mol |
| Purity: | >=98% |
| Type of Compound: | Alkaloids |
| Physical Desc.: | Powder |
| Source: | |
| Solvent: | Chloroform, Dichloromethane, Ethyl Acetate, DMSO, Acetone, etc. |
| SMILES: | COc1ccc(cc1CO)c1ccc2c(n1)nc(nc2N1CCOCC1)N1C[C@H](C)O[C@@H](C1)C |
| Contact us | |
|---|---|
| First Name: | |
| Last Name: | |
| E-mail: | |
| Question: | |
| Description | KU-0063794 is a potent and specific mTOR inhibitor, inhibiting both the mTORC1 and mTORC2 complexes with IC50s of 10 nM. |
| Target | mTORC1:10 nM (IC50) mTORC2:10 nM (IC50) |
| In Vitro | Ku-0063794 is cell permeant, suppresses activation and hydrophobic motif phosphorylation of Akt, S6K and SGK, but not RSK (ribosomal S6 kinase), an AGC kinase not regulated by mTOR. Ku-0063794 also inhibits phosphorylation of the T-loop Thr308 residue of Akt phosphorylated by PDK1 (3-phosphoinositide-dependent protein kinase-1). Ku-0063794 induces a much greater dephosphorylation of the mTORC1 substrate 4E-BP1 (eukaryotic initiation factor 4E-binding protein 1) than rapamycin, even in mTORC2-deficient cells, suggesting a form of mTOR distinct from mTORC1, or mTORC2 phosphorylates 4E-BP1. Ku-0063794 also suppresses cell growth and induced a G1-cell-cycle arrest[1]. Ku0063794 does not alter nuclear phospho-Mst1-Thr-120 levels in LNCaP cell nuclei, whereas Ku0063794 or CCI-779 increases phospho-Mst1-Thr-120 levels in C4-2 cell nuclei[2]. The combination of GDC-0941 and KU0063794 inhibits the phosphorylation of 4EBP1 and S6 to a similar extent to that caused by single agent NVP-BEZ235 in HCT116, DLD1 and HT29 cell lines[3]. |
| Cell Assay | Cells are treated with KU-0063794 for 24, 48, and 72 hours, and the medium is changed every 24 hours with freshly dissolved KU-0063794. For the measurement of cell growth, cells are washed once with PBS, and fixed in 4% (v/v) paraformaldehyde in PBS for 15 minutes. After washing once with water, the cells are stained with 0.1% Crystal Violet in 10% ethanol for 20 minutes and washed three times with water. Crystal Violet is extracted from cells with 0.5 mL of 10% (v/v) ethanoic (acetic) acid for 20 minutes. The eluate is then diluted 1:10 in water and absorbance at 590 nm is quantified. For the assessment of cell cycle distribution, cells are harvested by trypsinization, washed once in PBS, and re-suspended in ice-cold aq. 70% (v/v) ethanol. Cells are washed twice in PBS plus 1% (w/v) BSA and stained for 20 minutes in PBS plus 0.1% (v/v) Triton X-100 containing 50 g/mL propidium iodide and 50 g/mL RNase A. The DNA content of cells is determined using a FACSCalibur flow cytometer and CellQuest software. Red fluorescence (585 nm) is acquired on a linear scale, and pulse width analysis is used to exclude doublets. Cell-cycle distribution is determined using FlowJo software. |
| Density | 1.2±0.1 g/cm3 |
| Boiling Point | 694.3±65.0 °C at 760 mmHg |
| Flash Point | 373.7±34.3 °C |
| Exact Mass | 465.237610 |
| PSA | 93.07000 |
| LogP | 0.27 |
| Vapour Pressure | 0.0±2.3 mmHg at 25°C |
| Storage condition | Store at RT |